In complex gene editing projects, time is rarely on your side. Whether you’re running a genome-wide knockout to uncover novel disease targets or juggling multiple sub-libraries across different cell lines, the clock starts ticking the moment your project begins—and every delay risks missed opportunities, wasted resources, or results arriving too late to matter.
For many researchers, the challenge isn’t just getting the science right—it’s compressing a workflow that can stretch over months into something lean, predictable, and reproducible. Common bottlenecks include slow sgRNA design, limited access to stable Cas9 cell lines, uneven library coverage, and long validation cycles that stall progress just before the finish line.
A well-designed CRISPR screen can do more than identify functional genes—it can reshape your project timeline. By integrating high-coverage, low-noise screening directly into your workflow, you can spot valuable targets earlier, streamline downstream validation, and reduce the risk of costly re-runs. This guide breaks down where acceleration is possible and how Ubigene’s platform makes it real.
Choosing the Right CRISPR Screen — and the Service to Execute It
A CRISPR screen systematically asks: Which genes matter most for the phenotype I care about? By introducing thousands of sgRNAs into a cell population—each targeting a specific gene—you can track which edits affect measurable traits, from drug resistance to cell survival.
Main strategies:
| Type | When to Use | Pros | Limitations |
|---|---|---|---|
| CRISPR-KO | Knockout of coding genes; drug resistance; synthetic lethality | High efficiency; single-vector workflow | Lethal for essential genes; off-target risk |
| CRISPRi | Studying essential genes, regulatory regions | No genome breakage; reversible | Requires dCas9 line; scalability limits |
| CRISPRa | Gain-of-function, promoter mapping | No genome breakage; overexpression | Delivery constraints; accessibility issues |
Acceleration tip: Choosing the right screening type at the outset avoids dead-end experiments—e.g., using CRISPRi for essential genes or CRISPRa for promoter mapping can save weeks of trial and error.
If your project requires a streamlined, end-to-end CRISPR gene editing workflow—from gRNA design and library preparation to downstream validation—click here to explore Ubigene’s CRISPR gene editing service.
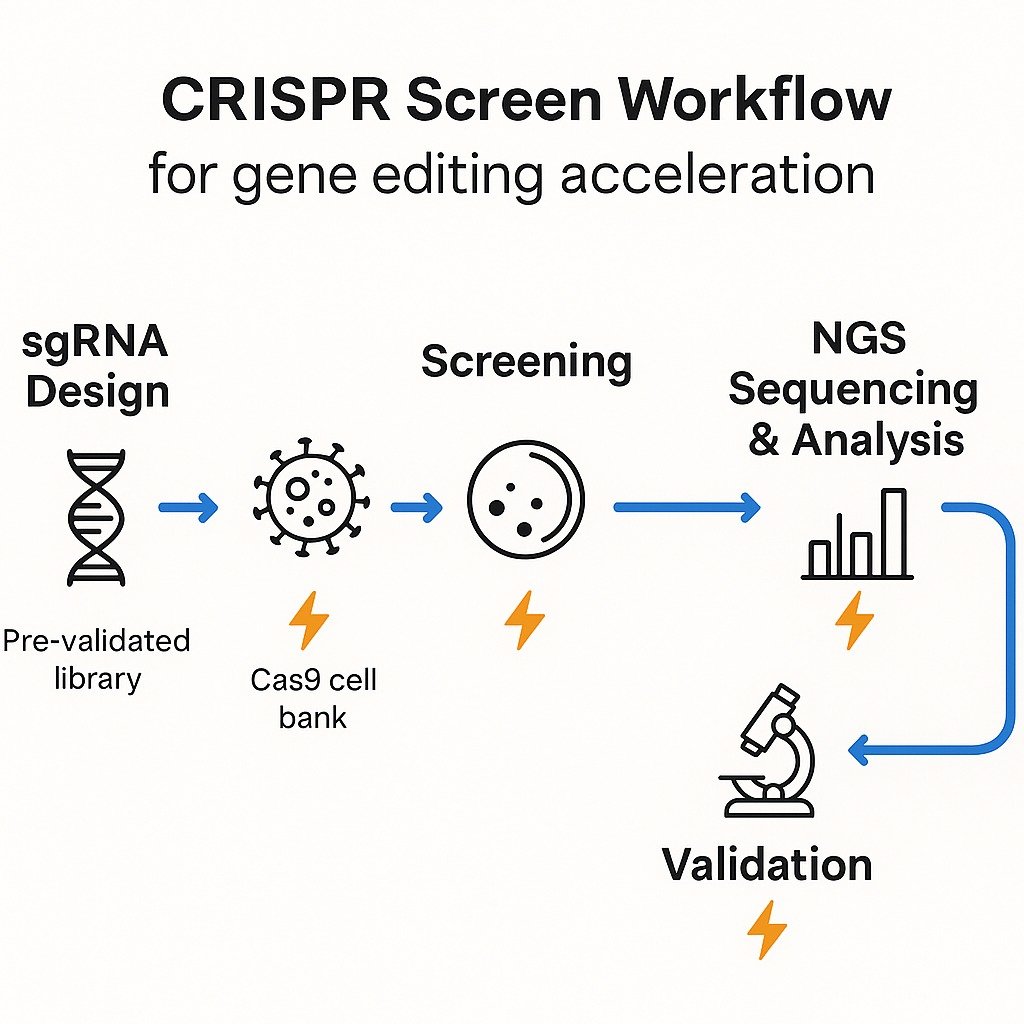
Where Time Is Won: Workflow with Acceleration Levers
While every lab has its nuances, CRISPR screening generally follows five stages. The fastest projects remove bottlenecks at each one.
- sgRNA Library Design & Construction
- Standard: 3–6 sgRNAs/gene, cloned into lentiviral backbones.
- Accelerate: Use automated scoring tools like Red Cotton™ for high-specificity designs; choose pre-built, validated libraries to skip redesign cycles.
- Watch out: Oversized libraries slow analysis—match scope to resources.
- Lentivirus Packaging & Transduction
- Standard: Package sgRNA pool, infect target cells at MOI <0.3.
- Accelerate: Start with Cas9-stable cell lines from a KO Cell Bank to skip 3–5 weeks; use high-titer, uniform viral prep to avoid repeat rounds.
- Watch out: Confirm titer/uniformity to prevent coverage loss.
- Screening Under Selective Pressure
- Standard: Apply drug, pathogen, or stressor.
- Accelerate: Pilot small-scale runs to calibrate selection strength; run controls in parallel to catch phenotype drift early.
- Watch out: Too harsh/mild pressure hides true hits.
- NGS Sequencing & Analysis
- Standard: Extract DNA, amplify sgRNAs, deep-sequence, analyze.
- Accelerate: Batch-process replicates in a single sequencing run; use preset QC thresholds in MAGeCKFlute + GSEA pipelines.
- Watch out: Low read depth forces costly resequencing.
- Candidate Validation
- Standard: Edit top hits individually; confirm phenotypes.
- Accelerate: Use pre-validated constructs; multiplex hits in one assay.
- Watch out: Always run rescue experiments to confirm causality.
How Much Time You Save with Acceleration
When every week counts, knowing exactly where time is saved can change how you plan your entire project. A conventional genome-wide CRISPR screen often takes 15–21 weeks, with major delays in library construction, Cas9 cell line generation, and downstream validation. By contrast, Ubigene’s accelerated workflow—featuring pre-validated libraries, ready-to-use Cas9-stable cell lines, high-uniformity lentivirus production, and integrated validation—can reduce total time to just 7–9 weeks. This time compression isn’t about skipping steps, but about optimizing them so you get high-quality, reproducible results faster. The chart below visualizes each stage and how these acceleration levers stack up.

Ubigene’s Acceleration Advantages
Many labs run CRISPR screens—few are built to accelerate them. Ubigene combines infrastructure, technical depth, and pre-validated resources to deliver speed without sacrificing data integrity.
- Red Cotton™ CRISPR Gene Editing Designer — Automated gRNA design with balanced cutting efficiency and low off-target risk.
- Library Coverage >99%, Uniformity <10%CV — Ensures every sgRNA has equal representation, avoiding missed hits.
- Cas9-Stable Cell Line Bank — Nearly 100 ready-to-use lines to skip time-intensive cell construction.
- High-Titer Lentivirus Production — Proven protocols for consistent, efficient transduction.
- Integrated Validation — Knockout, knockdown, or overexpression done in the same system—no tech transfer delays.
Impact in Real Projects
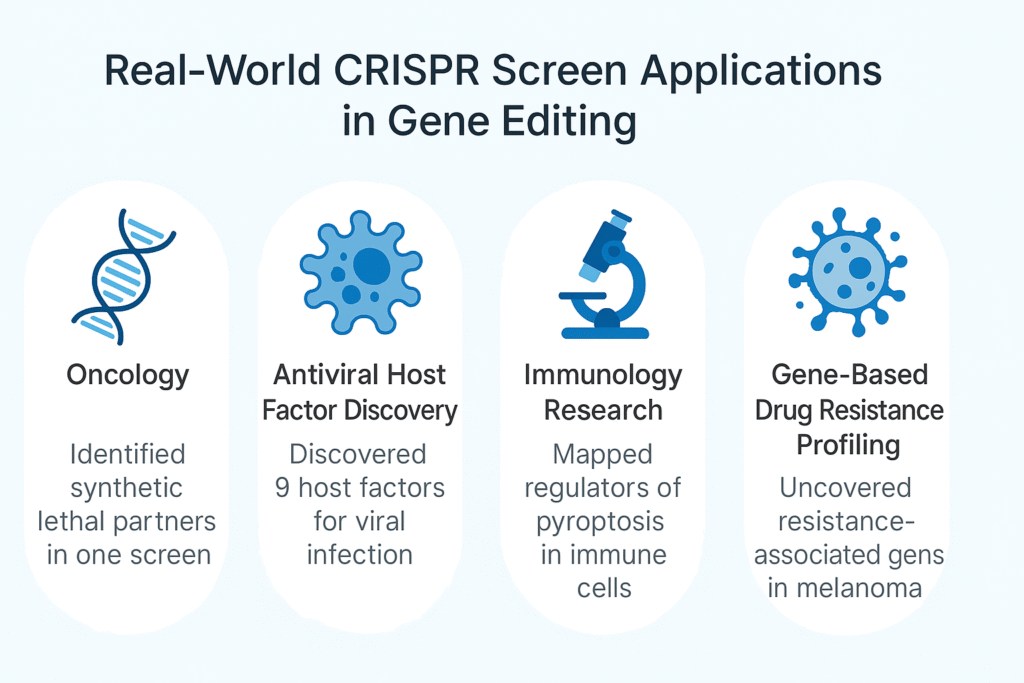
- Oncology Synthetic Lethality — Pre-validated Cas9 cancer lines cut prep by ~4 weeks; dual-gRNA KO library mapped lethal pairs in one cycle.
- Antiviral Host Factor Discovery — High-uniformity lentivirus maintained coverage; identified 9 ER-related factors without re-screening, saving 2 weeks.
- Immunology Pathway Mapping — Optimized selection + streamlined analysis condensed data processing from 3 weeks to 1.
- Drug Resistance Profiling — End-to-end workflow reduced total time from ~20 weeks to 9.
Cost Efficiency in Accelerated CRISPR Screening
Time saved is money saved—but the benefits go deeper. Cutting a project timeline from 20 weeks to 9 doesn’t just free calendar space, it reduces hidden costs and maximizes research ROI.
- Reduced Repeat Experiments — Pre-validated Cas9 lines, high-uniformity libraries, and streamlined CRISPR screen analysis lower the odds of re-runs.
- Lower Opportunity Cost — Faster completion can mean hitting grant deadlines or publishing ahead of competitors.
- Efficient Sequencing — Batching replicates in one high-throughput run reduces per-sample costs.
- Integration Saves Overhead — A single-vendor CRISPR workflow cuts shipping, QC duplication, and coordination delays.
- Predictable Budgets — Standardized accelerated workflows make funding requests and multi-screen planning easier.
Limitations & How to Handle Them
- Off-Target Effects — Validate with multiple sgRNAs and orthogonal methods.
- Delivery Constraints — Use pre-validated lines; test alternative delivery early.
- Complex Interactions — Start small with sub-libraries before scaling genome-wide.
- In Vivo Challenges — Use inducible/barcoded libraries; pair with in vitro validation.
- Data Bottlenecks — Standardize pipelines; automate QC to flag low-quality data early.
Future Directions in CRISPR Screen Acceleration
As the scale and complexity of biological questions continue to grow, the way we design and execute CRISPR screen projects is also evolving. One clear trend is the integration of AI-assisted gRNA design with multi-omics data, allowing researchers to prioritize targets based not only on genomic sequence but also on transcriptional and proteomic context. This shift will likely improve hit quality while further reducing the need for multiple screening cycles.
Another emerging direction is the use of fully automated, high-throughput CRISPR screening platforms, where library preparation, cell culture, and phenotypic readouts are handled in a closed-loop system. This approach minimizes human error, increases reproducibility, and accelerates turnaround from months to mere weeks.
Finally, combining CRISPR gene editing with spatial biology techniques and single-cell sequencing could unlock unprecedented insights—pinpointing not just which genes matter, but where and when they exert their effects in complex tissues. Ubigene’s adaptable workflows are positioned to integrate these advancements, ensuring that researchers can adopt cutting-edge methods without rebuilding their processes from scratch.
Conclusion
If your project demands both precision and pace, a CRISPR screen with built-in acceleration levers can be the difference between publishing first or playing catch-up. From gRNA design to final validation, Ubigene’s workflows are designed for speed, reproducibility, and data integrity.